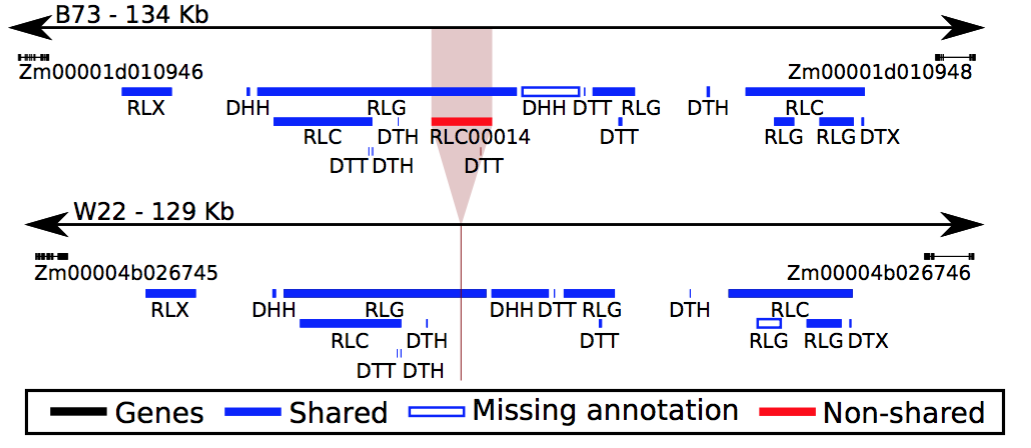
Transposable elements (TEs) are ubiquitous components of eukaryotic genomes and can create variation in genome organization and content. The majority of maize genomes are composed of TEs. We developed an approach to define shared and variable TE insertions across genome assemblies and applied this method to four maize genomes (B73, W22, Mo17, and PH207) with uniform structural annotations of TEs. Among these genomes we identified approximately 400,000 TEs that are polymorphic, encompassing 1.6 Gb of variable TE sequence. These polymorphic TEs include a combination of recent transposition events as well as deletions of olders TEs. There are examples of polymorphic TEs within each of the superfamilies of TEs and they are found distributed across the genome, including in regions of recent shared ancestry among individuals. There are many examples of polymorphic TEs within or near maize genes. In addition, there are 2,380 gene annotations in the B73 genome that are located within variable TEs, providing evidence for the role of TEs in contributing to the substantial differences in annotated gene content among these genotypes. TEs are highly variable in our survey of four temperate maize genomes, highlighting the major contribution of TEs in driving variation in genome organization and gene content.
Transposable elements contribute to dynamic genome content in maize
Anderson SN, Stitzer MC, Brohammer AB, Zhou P, Noshay JM, O'Connor CH, Hirsch CD, Ross-Ibarra J, Hirsch CN, Springer NM
2019.
The Plant Journal