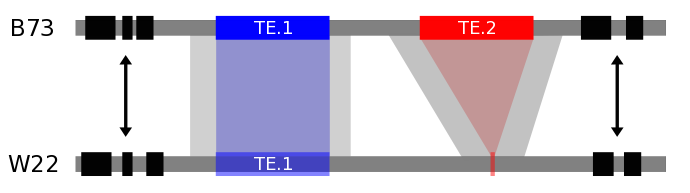
Transposable elements (TEs) constitute a large portion of crop genomes, and in maize, the majority of the genome is derived from LTR retrotransposons. TEs are highly variable across genotypes and make up a large amount of genetic variability across lines. We employ computational tools to assess which TEs are shared and variable across different genotypes in order to understand the causes and consequences of TE movement. As more assembled maize genomes become available, this will become an increasingly powerful tool for understanding the consequences of TE movement in maize.